2D and 3D Arabidopsis segmentation - BioImage.io
- jose-miguelserra-l
- Jan 1, 2016
- 2 min read
Updated: May 7, 2025
Authors
A. Wolny , Lorenzo Cerrone, Qin Yu
Description
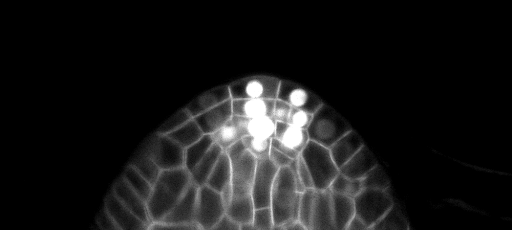
This is a set of models trained to predict the cell boundaries in confocal stacks of different parts of Arabidopsis : ovules, lateral root primordium and apical stem cells.
We have 2D models (slice by slice, smaller and faster models) and 3D models (slower but more accurate).
Input channel: confocal stacks, grayscale
Scale:
2D and 3D Unet Arabidopsis Ovules Cells, 0.235 µm x0.150 µm x 0.150 µm
3D Unet Arabidopsis Ovules Nuclei, 0.35 µm x0.1 µm x 0.1 µm
2D and 3D UNet Arabidopsis Apical Stem, 0.25 µm x0.25 µm x 0.25 µm
3D Unet Lateral Root Primordium, 0.25 µm x 0.1625 µm x 0.1625 µm
Bit depth: 8-bit or 16-bit
Output channel:
For cell boundaries: probability of cell boundaries
For nuclei: probability of nuclei
This model was downloaded and converted from BioImage.io, respecting the associated license (see License section below for more information)
Download
By downloading, installing, copying, accessing, or using the software, you agree to the terms of this end user license agreement.
Download model files with test images (ZIP archive)
Requirements
Make sure you have installed Aivia and the required DeepLearning module (according to our Wiki).
Installation and apply instructions
Aivia is required for applying the model file. You can request a demo copy of Aivia here.
Drag-and-drop the model file into the Recipe Console area; or use the 'Load recipe' option in the Recipe Console to load the model file.
Load the test image (or any image of your own) into Aivia.
If your image contains more than one channel, click on the 'Input & Output' section and specify the image channel you wish to apply the model on.
Click 'Start' to apply the model.
License
Copyright 2023, A.Wolny, L. Cerrone . MIT License. Full license information can be found here.
Acknowledgement
Bioimage.io is supported by AI4Life. AI4Life has received funding from the European Union's Horizon Europe research and innovation program under grant agreement number 101057970.
About AI4Life: https://ai4life.eurobioimaging.eu/.
References
Wolny, Cerrone et al. Accurate and Versatile 3D Segmentation of Plant Tissues at Cellular Resolution. eLife 2020. https://elifesciences.org/articles/57613







Comments